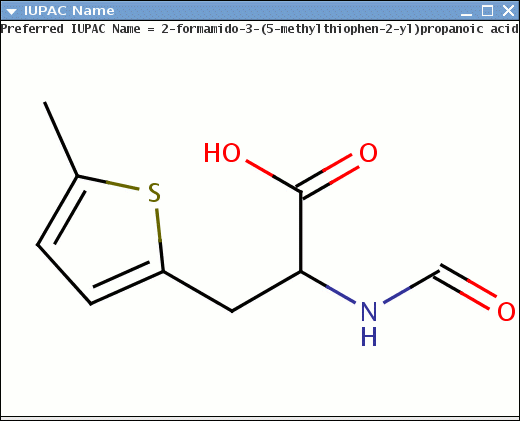

When possible, the generated name conforms to the IUPAC Provisional Recommendations for the Nomenclature of Organic Chemistry published in 2004. However, we do not claim full conformance with that document. Our current goal is to generate chemically correct names for as many cases as possible.
Importing IUPAC names is not supported yet, but it is under development.
ethane-1,2-diyl) are not supported yet.
With MarvinView, open the file containing the structures to be names. Then select the menu File/Save As, and choose "IUPAC Name files" in the "Files of type" drop-down box. Choose a name for the file, and click on the Save button. The file will contain the names of the structures, one per line.
Alternatively, on the command line, you can use the following command:
molconvert name inputs.mol -o names.txt
The file names.txt will contain the names of the molecules in the input file,
with one name per line.
It is possible to use a format option to chose a nomenclature style:
i (default) uses the IUPAC rules for prefered names;
t uses a more traditional style.
molconvert name:t inputs.mol -o names.txt
Adding names as an additional field to a SDfile can be achieved with the cxcalc tool.
cxcalc -S name input.sdf -o named.sdf